
Scholarly Git Experiences Part III: Quality Assurance
It has been a few months since I last posted, but the open research movement has not stopped even in today's pandemic. Let's start this post on a light note with yesterday's quarantine experience:

Now onto the good stuff. To date, I have discussed other features of these platforms, such as community building, education, and method tracking Part I and Part II. This post continues upon that work and focuses on quality assurance.
TL;DR previous posts, check out the chart below. Green rows highlight the items this post will focus on:
| Scholarly Experience Topics | Summary of Scholarly Experience Within a GHP | Related GHP Feature |
|---|---|---|
| Version Control | history of changes that allows scholars to gather a comprehensive understanding of how computations, scripts, and analyses have evolved through a study by viewing line-by-line changes in the codebase, as well as who in the research team they could contact for questions. |
|
| Community & collaboration | When researchers publish data, scripts, and software used in their study onto a GHP, there are lower barriers for reuse "and [it can] serve as a powerful catalyst to accelerate progress" (Ram, 2013). When readers and researchers have easy access to a resource, such as "sequence and metadata from microbiome projects", there's less of a chance to receive "missing, incomplete, inconsistent and/or incomprehensible sequence and metadata, and reluctance by authors, editors, and publishers to react to our complaints" (Langille, 2018). |
|
| Method tracking | Researchers are writing protocols to aggregate, manage, and analyze data into lab and field notebooks. Git commit history gives researchers and team members reference to understand the iterative progress of a study. Data analysis pipelines, also known as processing workflows, can benefit from version control systems for exposure, collaborative feedback, and reproducibility of the methodology. |
|
| Education | Syllabi, OER, and course management can be hosted on GHPs. As seen in scholarly research, students and teachers also benefit from a GHP as the platform enhances collaboration, communication, feedback, provenance of course materials and submitted work, and exposes students to industry experience. |
|
| Quality assurance | Merging small changes to the code often, throughout the development cycle. This tests bugs in data structure and code changes to ensure quality experimentation and analyses. |
|
| Reproducibility | NEXT | |
| Peer production | NEXT | |
| Publishing | NEXT | |
QA in Scholarship
QA == quality assurance. Commonly confused with quality control (QC), QA is a defined workflow that reveals whether or not "development and/or maintenance processes are adequate" for a system to meet standards and fulfill its purpose (The American Chemical Society). Meanwhile, QC is a process done after the final product is created in order to weed out or flag the unexpected. Various academic and commercial fields maintain standards that help ensure the systems building a product (from mechanical parts to software) meet a basic level of quality in terms of function, safety, style, format, accessibility, etc. For example, remember I Love Lucy and the chocolate factory line? The conveyor belt, machinery, cooking, and shaping the chocolate would need to undergo QA by a technical manager watching over the processes. One might argue that Lucy and Ethel eating the chocolates could be QC, but that's more of an unrelated operational issue that I won't address here.

We can look at web archiving workflows, an example that's close to home for team IASGE. Here, QA includes inspecting a captured webpage to ensure that all elements on the archived web page function properly and that URLs redirect to the correct pages. This allows researchers and other web archive visitors to have a smooth experience when looking into how geocities might have behaved or how HTML 1.0 would have rendered.
In the context of scholarly software development, QA steps are also taken before QC, but both the stages are necessary to ensure that features, functionality, and user experience of the software or script perform as expected with each new code release. QA takes place throughout the development process; it is like the flu shot, or practicing a consistent & healthy diet—preventing bugs from invading the system. The QA process flags low quality outputs and analyses as well as code errors, allowing developers to address and fix bugs before sharing with collaborators and fellow researchers (Widder, et al., 2019; Yenni, et al., 2018).
Continuous Integration (CI)
CI is a practice or workflow included into software development workflows. CI platforms that facilitate the workflow are like robots that integrate with developing environments and "tell you, every time you change your code, if it still works or if you broke it" (Nowogrodzki, 2019), which can be likened to QA processes. Each integration of code can then be verified by automated builds and tests written by the developers. CI services can come from an external third-party system, but GitHub Actions and GitLab CI are popular services native to their respective GHP. Other popular CI platform options that integrate with GHPs and have been cited in scholarly research are Travis CI, CircleCI, and Jenkins (Bendetto, et al., 2019). See awesome-ci for a more robust list of CI services. Many of these are proprietary platforms, but similar to GHPs, they rely on the open source community as their gateway user base to build into their premium subscription business model. Travis CI is the most popular CI service and has even been touted by the Zoteroist as the automated first wall of defense.
To begin with CI, you must write a YAML file (specific to your CI service) in the Git repository that outlines the dependencies, tests, and workflows for running your analysis or data processing scripts (e.g. what comes first—preferably, the dependencies!). When new changes are pushed to that repository, the CI service reads the YAML file to build the test environment and run the steps as you specify them. While each YAML files' structure will differ depending on specific workflows and needs, here's a breakdown of how the IASGE YAML file that builds our website with GitLab CI:
image: registry.gitlab.com/paddy-hack/nikola:8.0.1This first line in the YAML file identifies which Docker container we want to use from the GitLab Docker registry, which ensures we are starting with all the computational dependencies we need.
test:
script:
- nikola build
except:
- mastertest: initiates the building of the test environment so that the newly added code can be tested before going to a production stage.
pages:
script:
- pip3 install pybtex
- nikola plugin -i publication_list
- nikola build
artifacts:
paths:
- public
only:
- masterpages: installs the extra dependencies that we need (the publication_list plugin and its dependency, pybtex) and then tries to build the website with the relevant Nikola commands. If it succeeds, it will publish the new changes to the website. If it fails, we are notified and the changes do not proceed.
CI for Scholarly Code
When developing and using software, scripts, or code for experiments or analyses, scholars can integrate QA into their research processes through "the practice of merging in small code changes frequently—rather than merging in a large change at the end of a development cycle" (Travis CI documentation, n.d.). This practice is also known as continuous integration (CI). As research is becoming reliant on (a) correctly structured and processed data, and (b) increasingly homegrown software, pursuing QA for both (data and software) through CI is one way to automate both—checking that data is well-structured and that the code works the way it is expected to after those many small merges.
Since CI is commonly integrated with code repositories (and in this case git repos), this allows the CI configuration files (used to build environments, tests, and pipelines) to be edited and used by multiple people, while performing QA on snippets of code on a remote server. Bugs are flagged before versions are merged and integrated into the master codebase, helping researchers catch these early and often (GitLab Continuous Integration (CI) & Continuous Delivery (CD)). A well-documented workflow starting from scratch is an excellent approach for improving reproducibility and replicability efforts (to be detailed in my next post).
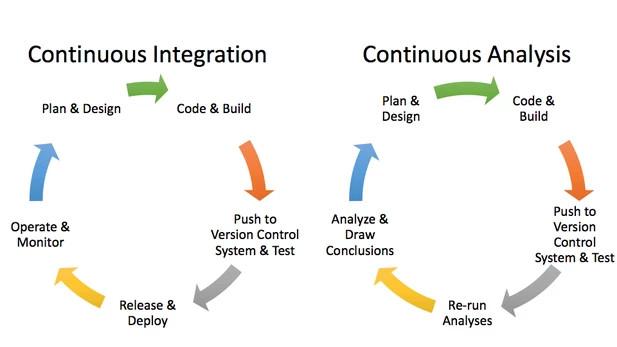
However, CI for QA is useless without robust tests in place. While software engineering relies on testing processes to verify consistency within their QA checkpoints, this mirrors the term "scientific testing" redefined by Krafczyk, et al. (2019). In this case, scientific testing benefits from black-box (aka system) testing for being able to reproduce the computational environments and results being published in articles, as opposed to white-box (aka unit) testing. White-box testing is a QA practice that looks to verify only "specific functions or sections of code are [performing] properly". By contrast, black-box strives "to test [the behavior of] the whole application" (Krafczyk, et al., 2019), also referred to as specification-based testing. The tester (CI, in this case!) is aware of what the software is supposed to do but is not aware of how it does it. Here's one example: say that your research relies on merging together lots of different spreadsheets. You know that you are expecting one ID column from each spreadsheet, which is a number between 0000-1000. In the CI configuration file, you can write a test such that if there is any ID number outside that range, or if any values in that column are not numbers, it fails.
Krafczyk, et al. (2019) sought to "construct testing scripts capable of running" whole computational procedures based on existing public scholarly repositories on GitHub, in which they found testing "minimized [versions of] computational experiments" are less time consuming and can accurately "mimic the original work... at a smaller scale" (Krafczyk, et al., 2019). Benefits from this minimized version testing approach include flagging software issues when changes are made, as well as identifying if "the results are sensitive to the computation environment" (Krafczyk, et al., 2019).
Andreas Mueller, Associate Research Scientist at Columbia University's Data Science Institute, discusses that with CI, researchers writing and running code to perform experiments or analyze data can be tested by "each individual component of the software to ensure that it is sound." In implementing CI, researchers and developers are able to:
- streamline and avoid tedious, manual work that is susceptible to errors
- make incremental code changes, lowering the risk of significant bugs or breaks
- receive fast review and feedback on code updates, allowing researchers to address bug fixes immediately and understand next steps needed for procedures and data collecting.
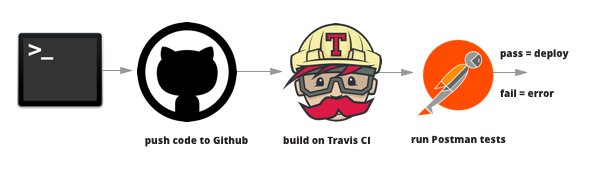
For example, the Fermi National Accelerator Laboratory has written about their own CI service setup, where lab members collaborating with data collection and analytical code. It was clear (1) that the data needed to be properly structured, and (2) code updates were compatible through processing the data for analyses such as "comprehensive testing from unit tests to physics validation through regression test on multiple platforms/compiler/optimization that are supported by code developers" (Bendetto, et al., 2019). They are able to implement tests on code pull requests by implementing their own CI service with a Jenkins set up, which executes these phases sequentially: "environment setup, code checkout, build, unit tests, install, regression tests and validation tests" (Bendetto, et al., 2019). With the CI validation tests, researchers are able to monitor "the working directory; initialization and finalization procedures; the command to execute the task; logs to report; how to filter logs sent by the final report" (Bendetto, et al., 2019).
Another example comes from the University of Florida's Department of Wildlife Ecology and Conservation who set up "R code to error-check new data and update tables" (Yenni, et al., 2018) through Travis-CI implemented on their GitHub repo. CI will push notifications if changes to the code breaks the workflow, allowing researchers to immediately address the issue(s). It's like having a collaborator manually test the code experiment, and then send you an issue request with an error message if they were unable to replicate your code to expected results.
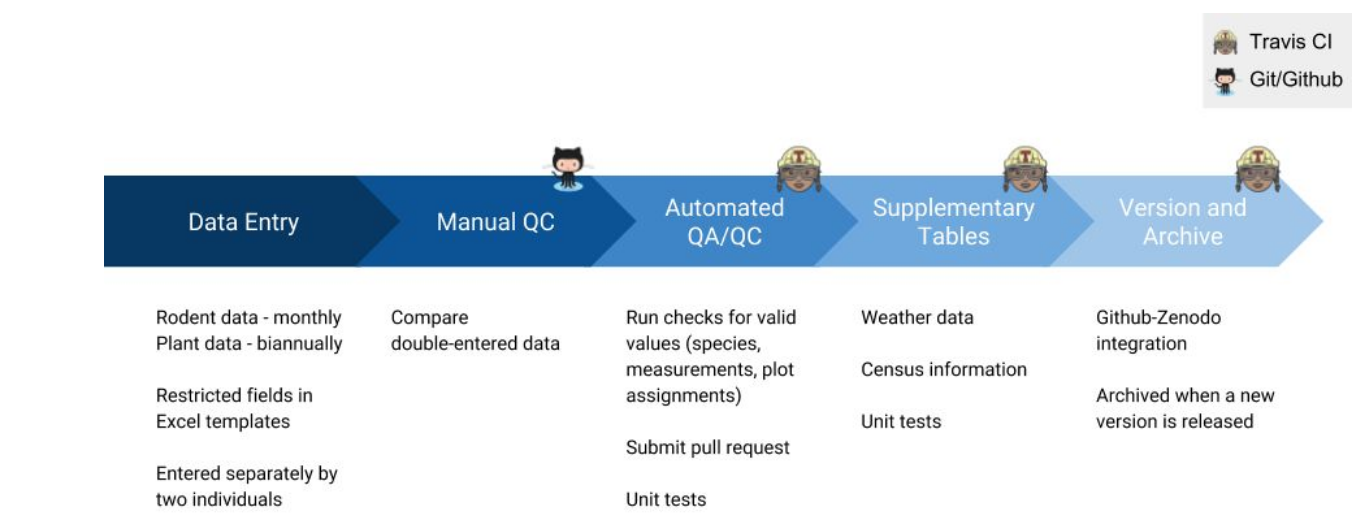
In short, this is the CI process within research:
- Researcher builds software or script and pushes it to update the Git repository.
- CI program clones the Git repo into a new and separate cloud-based virtual environment, usually defined by a Docker container and additional dependency installations.
- Code builds, runs, and tests in that new virtual environment through a series of tasks defined in a configuration file.
- CI will detect failed and successful tasks, and will send notification if the code is broken or passed, respectfully (Travis CI documentation, n.d.).
Et voilà, code can be updated to ensure its quality is up to standards and researchers can spend more time on innovation!
Issues with CI in Scholarship
And last, nothing is perfect in technology nor life, so here are some of the cons to CI with GHPs for scholars and researchers. Widder, et al. (2019) does a great job in providing a comprehensive review of the known issues in adopting and/or using CI workflows and services within software development teams. From a full literature review, Widder, et al. (2019) categorizes five major pain points in CI within software developing workflows: information overload, organizational culture, setup and configuration, slow feedback, and testing deficiencies. Luckily, Krafczyk, et al. (2019) has proven examples in how to address CI’s slow feedback challenges through minimized version testing, as mentioned in the beginning of this post. Through mixed methods research (e.g. survey for quantitative and interviews for qualitative), Widder, et al. (2019) published a thorough table (4: Results Summary) of their findings in what pain points of CI could increase or decrease the likelihood of adopting the workflow and/or switching to incorporate CI. A few categories include: lack of support for specific languages from certain CI services, infrequent changes to the code itself means less of a need to constantly push tests and validation, and inconsistent CI practices among team members makes for a messier codebase. But, don’t let this deter you from trying out CI to help automate some QA workflows for your team! Giving it a try can yield really helpful, time-saving results, as outlined above.
Conclusion
In the end, CI can enable researchers to make progress with experiments, uninterrupted, unless an issue pops up from a CI run. While it's built for coders, scholars and researchers can benefit from the use of CI to automate (and validate!) parts of folks' research workflows. It is an interesting application of GHPs within a research context, and we look forward to seeing more CI in research repos as we move forward in our IASGE research.
As mentioned in previous posts, these IASGE updates are active conversations while I continue my research about the scholarly Git experience. We encourage questions and further research suggestions. Please feel free to submit an issue or merge request to our GitLab repository or email Vicky Steeves and/or me to continue the conversation. Thanks for tuning in!
Bibliography
Akhmerov, A., Cruz, M., Drost, N., Hof, C., Knapen, T., Kuzak, M., Martinez-Ortiz, C., Turkyilmaz-van der Velden, Y., & van Werkhoven, B. (2019). Raising the Profile of Research Software. Zenodo. https://doi.org/10.5281/zenodo.3378572
Alexander, H., Johnson, L. K., & Brown, T. (2018). Keeping it light: (re)analyzing community-wide datasets without major infrastructure. GigaScience, 8(2). https://doi.org/10.1093/gigascience/giy159
Bai, X., Pei, D., Li, M., & Li, S. (2018). The DevOps Lab Platform for Managing Diversified Projects in Educating Agile Software Engineering. 2018 IEEE Frontiers in Education Conference (FIE), 1–5. https://doi.org/10.1109/FIE.2018.8658817
Beaulieu-Jones, B. K., & Greene, C. S. (2017). Reproducibility of computational workflows is automated using continuous analysis. Nature Biotechnology, 35(4), 342–346. https://doi.org/10.1038/nbt.3780
Benedetto, V. D., Podstavkov, V., Fattoruso, M., & Coimbra, B. (2019). Continuous Integration service at Fermilab. EPJ Web of Conferences, 214, 05009. https://doi.org/10.1051/epjconf/201921405009
Krafczyk, M., Shi, A., Bhaskar, A., Marinov, D., & Stodden, V. (2019, June 1). Scientific Tests and Continuous Integration Strategies to Enhance Reproducibility in the Scientific Software Context. 2nd International Workshop on Practical Reproducible Evaluation of Computer Systems (P-RECS’19). ACM Federated Computing Research Conference, Phoenix, Arizona. https://doi.org/10.1145/3322790.3330595
Stolberg, S. (2009). Enabling Agile Testing through Continuous Integration. 2009 Agile Conference, 369–374. https://doi.org/10.1109/AGILE.2009.16
Widder, D. G., Hilton, M., Kästner, C., & Vasilescu, B. (2019). A conceptual replication of continuous integration pain points in the context of Travis CI. Proceedings of the 2019 27th ACM Joint Meeting on European Software Engineering Conference and Symposium on the Foundations of Software Engineering, 647–658. https://doi.org/10.1145/3338906.3338922
Yenni, G. M., Christensen, E. M., Bledsoe, E. K., Supp, S. R., Diaz, R. M., White, E. P., & Ernest, S. K. M. (2018). Developing a modern data workflow for evolving data. BioRxiv, 344804. https://doi.org/10.1101/344804