
Scholarly version control, community, & method tracking with Git hosting platforms (GHPs)
Even when there are illustrative zines and guides in layman's terms about Git and Git hosting platforms (re: Oh shit, git!, Really Friendly Git Intro, or Non-technical person's guide to becoming an open source software contributor via Github), it is still difficult for many overcommitted scholars to re-imagine their research habits. Many still see a barrier to entry when incorporating a git commit and a git push at the end of the day of doing their research. As with many first impressions, it's hard to shake off the idea that Git was originally created by and for computer scientists. There will also always be a collaborator who will say "screw it, can we just use Dropbox?" (Raj 2016). This is all true, but as more users adopt Git hosting platforms (GHPs), new features are developed or hacked to create more relevant scholarly features. This post will cover four of the eight (and counting) notable ways that scholars have incorporated GHPs, specifically GitHub, GitLab, and BitBucket, into their daily research, documentation, collaboration, application, and advocacy for openness: (1) version control, (2) community & collaboration, and (3) method/protocol tracking.
1. Version control
Version control, also known as revision-control or source control, is the foundation to Git's existence, but its functionalities become even more powerful with a GHP in-play. In Git, each version is marked with a revision number to distinguish records. Thus, "every copy of a Git repository carries a complete history of all changes, including authorship, that can be viewed and searched by anyone. This feature allows new authors to build from any stage of a versioned project." (Ram 2013) The staged version can then be assigned to a peer for review using the issue tracker or pull request features available on Bitbucket, GitHub, and GitLab before committing to updating the master file (or branch). Informative and well-written comments from contributors, can make revisions easier to address into newer versions. These can be incorporated into larger project management interface (e.g. Jira, Trello, Basecamp) where issues can be assigned to individuals, prioritized by voting, and can be bolstered with other productivity features such as dates, milestone, and metadata categorization.
There are a couple options for reviewers to survey code revisions to understand the changes done in a file. Similar to word processing document track changes, GHPs can display the commit change logs stored in Git (also known as history timeline or commit history), either in-line or side-by-side views that highlight deletions and additions.
In command-line, these version logs are prompted by git log or git diff.
If a researcher is keeping their local Git repository up-to-date via (git push and git pull), then the researcher will not only be able to view the changes they've made themselves, but they will also be able to see how their peers have altered the file with new findings or ideas.
This granular version control metadata allows scholars to gather a more comprehensive understanding on how computations, scripts, and analyses have evolved through a study by viewing line-by-line changes in the codebase, as well as who in the research team they could contact for questions.
2. Community & collaboration
Good scholarship includes open sharing of teaching, research, and service materials to peers within local and international communities, as well as across disciplines. The willingness to be open and transparent about methods, data, and code is crucial to prompting opportunities for further research. GHPs can fulfill these type of community contributions.
When researchers publish data, scripts, and software used in their study onto a GHP, there are lower barriers for reuse "and [it can] serve as a powerful catalyst to accelerate progress." (Ram 2013) When readers and researchers have easy access to a resource, such as "sequence and metadata from microbiome projects", there's less of a chance to receive "missing, incomplete, inconsistent and/or incomprehensible sequence and metadata, and reluctance by authors, editors, and publishers to react to our complaints." (Langille 2018) With codebase and data published on a GHP, readers will have more agency to expand ideas and conduct their hypotheses within consistent "software versions [and] program parameters" (Langille 2018) as they reference the original publication. Similar to a travel visa, my Vietnamese cousin can only travel to fifty-four foreign countries without a visa. Because of the extra paperwork for travel approval, she does not get to travel with ease she is limited to progress in exploring foreign cultures around the world. On the other hand, as a U.S. citizen, I am able to travel to 166 countries without a visa. Without the bureaucratic paperwork, my ability to experience foreign lands is much more attainable than my cousin's. When a research paper states "data is available upon request" (Langille 2018), this requires readers to first find the author's contact information, reach out to the author in hopes that the contact information is current—a study on disappearing research data with articles' age by Vines, Albert, et al. notes that finding a "working email address for the first, last, or corresponding author fell by 7% per year." (Vines 2013). If luck is on our side and we don't receive a Mailer Daemon reply, then the author must in return, be willing to take time to respond and manually share with each individual reader request. These hurdles slow down the process of access and execution of new ideas based on original data. Creating an open community of sharing data will enhance efforts to reference and boost each others' works.
Scholars can import code and data onto a GHP. Bitbucket, GitHub, and Gitlab all offer simple upload buttons on their GUI web browser, along with a drag-and-drop file option. There is variety in the design and location of each platform's upload button, but they are all point-and-click.
The command-line version is exactly the same for all three platforms, using the common Git commands after creating the file:
git add <files>
git commit -m "[message]"
git push
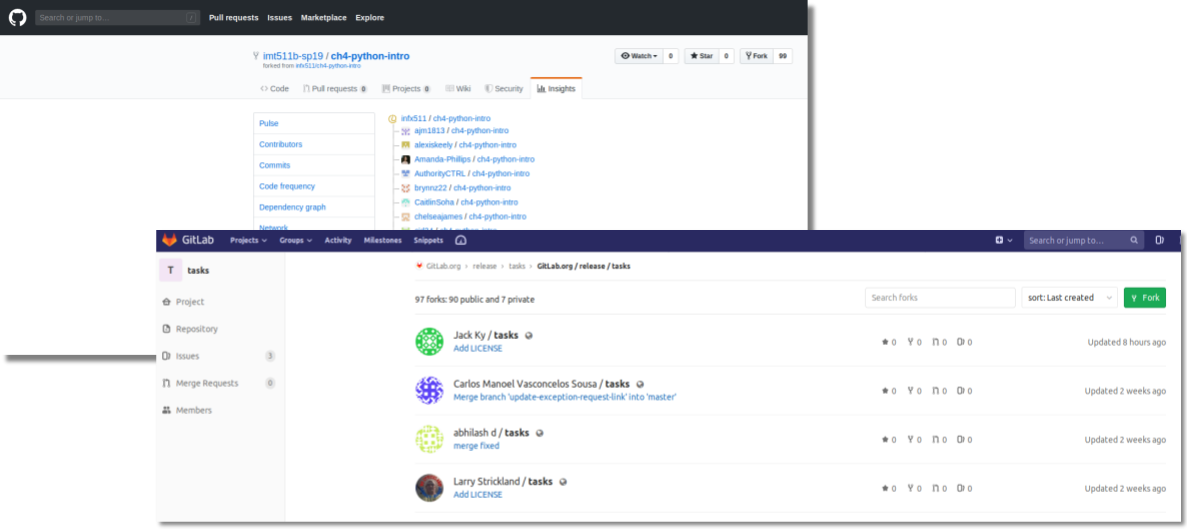
Once codebase is shared on the GHP, readers will be able to take advantage of the platform's features to not only reuse the codebase through forking, as well as use features like bug reporting, issue tracking, and viewing authorship and comment logs to communicate with the author and fully understand the study. The list of forks, a network, is another way for authors to see who is referencing their works and how it is being remixed.
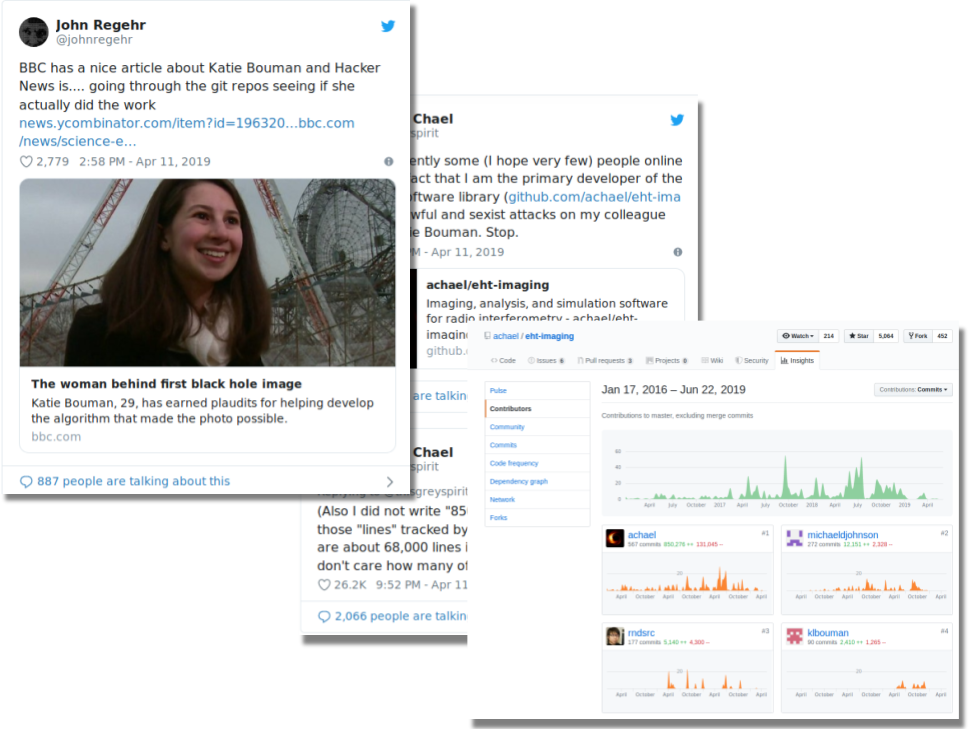
Bug reporting and issue tracking can be tools for collaboration on lab notebooks, similar to a Google Doc. In publishing, "git commit logs can provide a highly granular way to track and assess individual author contributions to a project.. [and] commits are attributed to specific authors, error or clarifications can also be appropriately directed." (Ram 2013) While these quantitative measures can't be the end-all-be-all in looking at authorship, they can spark collaborations and networking between authors and researchers who would not have known each other's body of work, giving appropriate attribution to the correct person. It also supports the movement of "high profile journals now discouraging the practice of honorary authorship." (Ram 2013)
A great example is from the University of Washington's Director of Open Software, Jake Vanderplas' GitHub repository Mutiband Lomb-Scargle Periodograms (multiband_LS). In 2014, he and a collaborator wrote this research paper in GitHub from the beginning ideation phase to the fully published paper. We can see in 2015, when the paper was most likely almost finished, they received an unsolicited-but-helpful issue —Scope of Jaynes analysis/Bayesian formalism #1—from Jeff Scargle of NASA Ames Research Center, who Vanderplas et. al. cite in their paper. The conversation on the issue ticket is informative, honest, and helpful to Vanderplas' research. This type of peer review made me think twice about the benefits of the hard-driven anonymous peer review process that is usually managed in the dark by publishing companies. It makes me think there's a chance with transparent GHPs, that there is opportunity to skip the intermediary predatory publishers, and make discussion more open and available for readers to understand how and why a conclusion came to be.
3. Method/protocol tracking
Researchers are writing protocols to aggregate, manage, and analyze data into proper lab and field notebooks. Normally, these records are recorded into the lab/research notebook that the team should be able to reference in order to understand the progress of the study. This includes hypotheses, observations, reflections, and analyses. Benchling and protocols.io are common proprietary-freemium lab notebook platform. On the other hand, there's open source Git and GHPs, where each change and push to update code or files requires a commit message. A good commit message is written as a present tense statement, detailed enough to communicate changes without having to look at the code, and 50-72 characters long. (Oliviera 2019) These "Git commit logs can serve as proxies for lab notebooks… over the course of a project", and for scientists, this is useful since "every copy of a repository carries a complete history of changes available for anyone to review." (Ram 2013)

General data analysis pipeline, also known as processing workflows, benefits from version control systems as well. For example, Arjun Raj's systems biology lab, used Mercurial and Bitbucket to "refactor [their] image analysis code… to adopt version control for these general image processing tools." (Raj 2016) At the time, the lab's researchers were proficient in scripting and computation so they saw opportunity in using Bitbucket to host "various analysis scripts that will take this data, perform whatever statistical analysis and so forth on it, and then turn that into a graphical element. Typically, this is done by one, more often two, people in the lab, typically working closely together."(Raj 2016) They also mention that there is constant turnover rate for lab grad students and researchers, as is natural in academia, which also changed the proficiency-level and tendency towards Git and GHP workflows.
While many believe in the opportunities that the "commit history could serve as a computational lab notebook," there will always be hesitant adopters. In a blog post, Raj also states that "version control is not a notebook, keylogging is not a notebook, the only thing that is a notebook is you actually spending the time to write down what you did, carefully and clearly–just like in the lab." (Raj 2016) Depending on the operational dynamics, existing skills, and willingness to learn, the Git commit message logs have the potential to supplement and structure time-consuming lab notebooks.
On the other hand, there are others who have found success in hosting lab notebooks on Github—Tad Dallas' Lab Notebook from the Centre for Ecological Change at the University of Helsinki, the protein-systemic-characterization lab notebook from the Novartis Institutes for Biomedical Research, and others who use GitHub Pages and Jekyll as a simple log to host their field notes (e.g. aquatic and fishery sciences lab notebook). There is also open source lab software like eLabFTW, which you can fork, download, and host on your own.
The scholarly experience has embraced software and digital interfaces as primary tools to conduct, gather, and disseminate works. More researchers incorporate software, scripts, programs, or computations into their subjects and analyses, and so it is only natural that we need to find better ways to ensure these digital artifacts are able to sustain its true form across time and for the diversity of potential users. These Git hosting platforms offer similar features that enhance the domains that define the scholarly experience community building, methodology bread crumbs, and version control. In a few weeks, I'll be able to share more findings within Git-scholarly methodologies.
Through written works, ranging from formal journal article publications to informal documentation blogs, it is clear that academics have jumped on the software development bandwagon and are taking advantage of platforms like GitHub, GitLab, Bitbucket, and SourceForge. As the scholarly and pedagogical practices around Git are still new, this project, Investigating & Archiving the Scholarly Git Experience (IASGE), looks forward to sharing our quantitative and qualitative data and analyses on how GHPs truly affect the scholarly experience. In the coming months, questions can be clarified through further research such as scraping of academic journal databases with articles referencing to a Git hosting platform, a survey gathering quantitative data on usage practices, and qualitative interviews with select users.
Bibliography
Langille, M. G. I., Ravel, J., & Fricke, W. F. (2018). "Available upon request": not good enough for microbiome data! Microbiome Journal, 6(8). https://doi.org/10.1186/s40168-017-0394-z
Ma, E. J. (2016). GitHub Lab Notebooks. Retrieved July 1, 2019, from ericmjl website: https://ericmjl.github.io/blog/2016/9/6/github-lab-notebooks/
Raj, A. (2016b, March 3). From over-reproducibility to a reproducibility wish-list [Blog]. Retrieved April 25, 2019, from RajLab website: http://rajlaboratory.blogspot.com/2016/03/from-over-reproducibility-to.html
Ram, K. (2013). Git can facilitate greater reproducibility and increased transparency in science. Source Code for Biology and Medicine, 8(7), 8. https://doi.org/10.1186/1751-0473-8-7
Vanderplas, J. (2016, April). Driving Reproducibility at UW. Presentation presented at the NYU Reproducibility Symposium, Brooklyn, NY.
Vines, T. H., Albert, A. Y. K., Andrew, R. L., Débarre, F., Bock, D. G., Franklin, M. T., … Rennison, D. J. (2013). The Availability of Research Data Declines Rapidly with Article Age. Current Biology, 24(1), 94–97. https://doi.org/10.1016/j.cub.2013.11.014